Introduction
I am using waterfall charts drawn in ggplot2 to visualize GLM coefficients, for regression and classification. Source Rmd file can be found here.
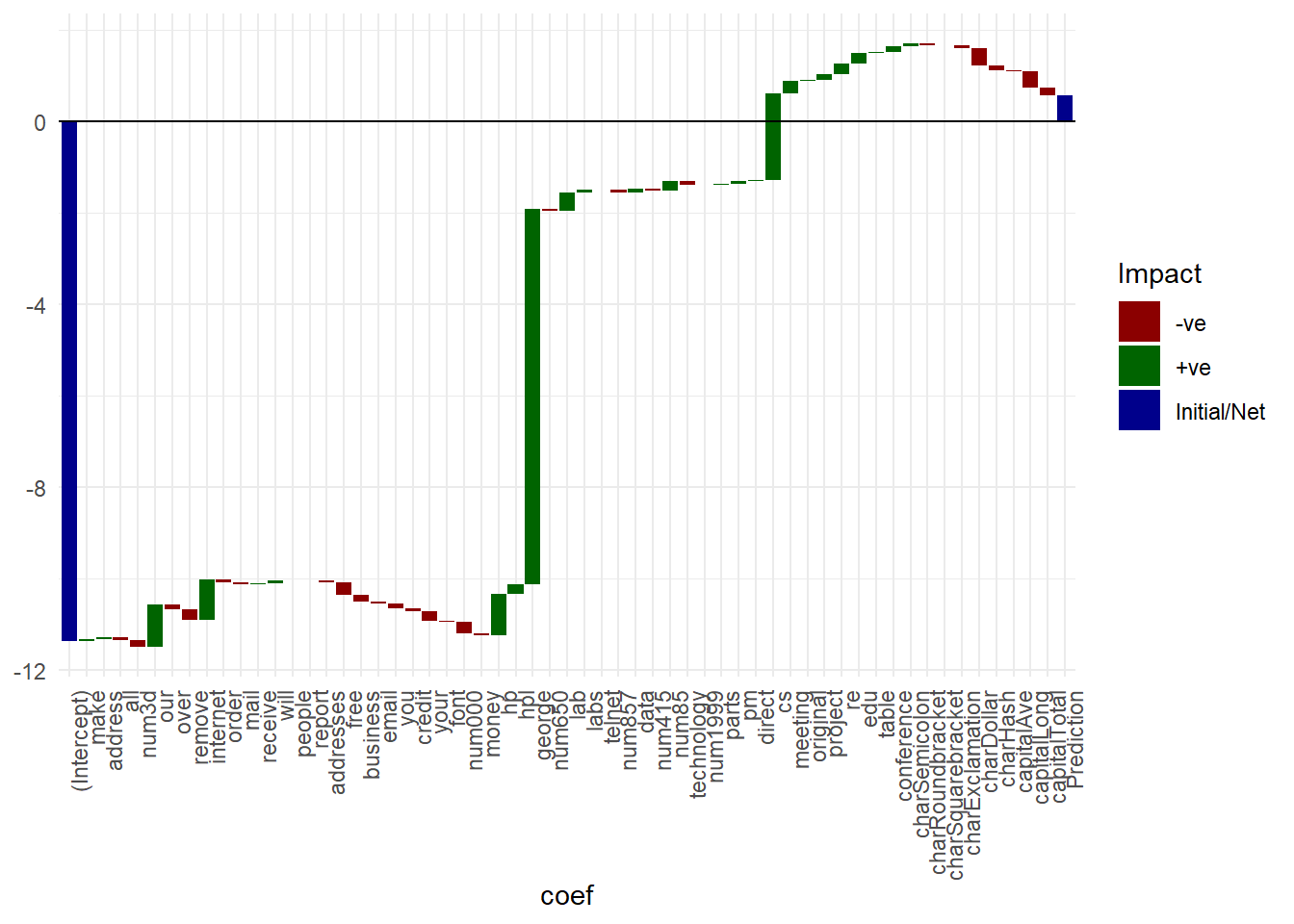
- Waterfall chart: inspired by their commonplace use in finance1, a simple visualization to illustrate the constituent components (numeric values) that make up the final model prediction, starting from the intercept term . The idea is quickly see which features contribute positively and which negatively, and by how much. Important thing to note here is that the waterfall chart will differ from test datapoint to test datapoint - we first have to make a prediction using a test sample , get the prediction, then visualize individual absolute feature contribution to the prediction .

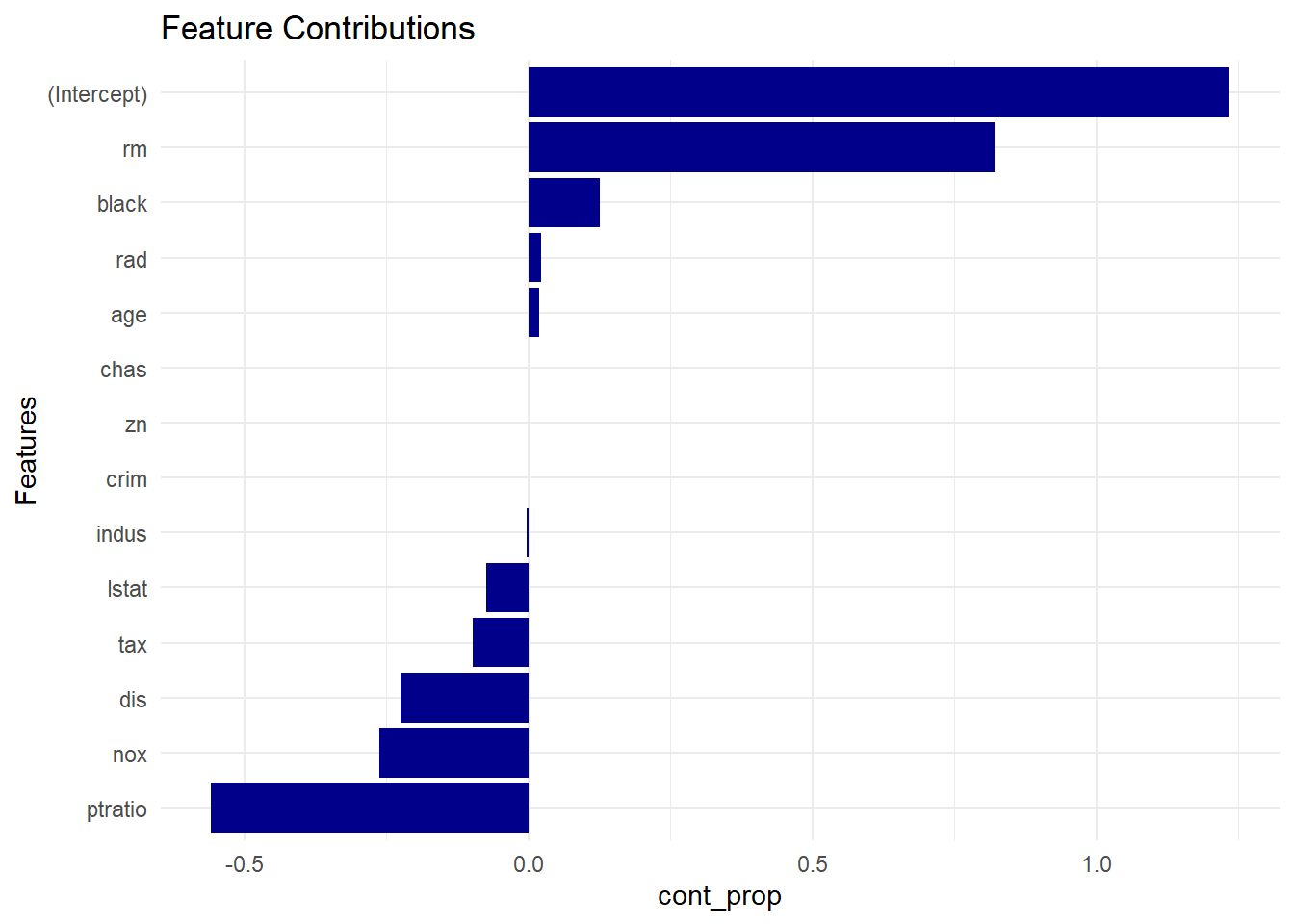
- Feature contributions chart: this one is simpler. Same idea as above (also dependent on test sample ), but plotted by ranking the numeric contributions by their proportions relative to the prediction2 like this:
contribution_proportion = feature_contribution / prediction, written below ascont_prop <- featcont/pred.
Regression
Preparation
library(MASS)
library(caret)
## Warning: package 'caret' was built under R version 3.6.3
## Loading required package: lattice
## Loading required package: ggplot2
## Warning: package 'ggplot2' was built under R version 3.6.3
library(magrittr)
library(ggplot2)
data(Boston)
set.seed(123)
# mean centering
b2 <- preProcess(Boston, method = "center") %>% predict(., Boston)
idx <- createDataPartition(b2$medv, p = 0.8, list = FALSE)
train <- Boston[idx,]
test <- Boston[-idx,]
mod0 <- lm(data = train, medv ~.)
sm <- summary(mod0)
betas <- sm$coefficients[,1]
testcase <- test[1,]
pred <- predict(mod0, testcase)
# dot product between feature vector and beta
featvec <- testcase[-which(testcase %>% names == "medv")] %>% as.matrix
betas2 <- betas[-1]
nm <- names(betas)
#betas2 %*% t(featvec)
# feature contributions
featcont <- betas2*featvec
featcont <- c(betas[1], featcont, pred)
names(featcont) <- c(nm, "Prediction")Waterfall chart on regression feature contributions
# waterfall chart on feature contribution
plotdata <- data.frame(coef = names(featcont), featcont = featcont, row.names = NULL)
plotdata$coef <- factor(plotdata$coef, levels = plotdata$coef)
plotdata$id <- seq_along(plotdata$coef)
plotdata$Impact <- ifelse(plotdata$featcont > 0, "+ve", "-ve")
plotdata[plotdata$coef %in% c("(Intercept)", "Prediction"), "Impact"] <- "Initial/Net"
plotdata$end <- cumsum(plotdata$featcont)
plotdata$end <- c(head(plotdata$end, -1), 0)
plotdata$start <- c(0, head(plotdata$end, -1))
plotdata <- plotdata[, c(3, 1, 4, 6, 5, 2)]
gg <- ggplot(plotdata, aes(fill = Impact)) +
geom_rect(aes(coef,
xmin = id - 0.45,
xmax = id + 0.45,
ymin = end,
ymax = start)) +
theme_minimal() +
#scale_fill_manual(values=c("#999999", "#E69F00", "#56B4E9"))
scale_fill_manual(values=c("darkred", "darkgreen", "darkblue")) +
theme(axis.text.x=element_text(angle=90, hjust=1))
## Warning: Ignoring unknown aesthetics: x
#coord_flip()
if(sign(plotdata$end[1]) != sign(plotdata$start[nrow(plotdata)]))
gg <- gg + geom_hline(yintercept = 0)
gg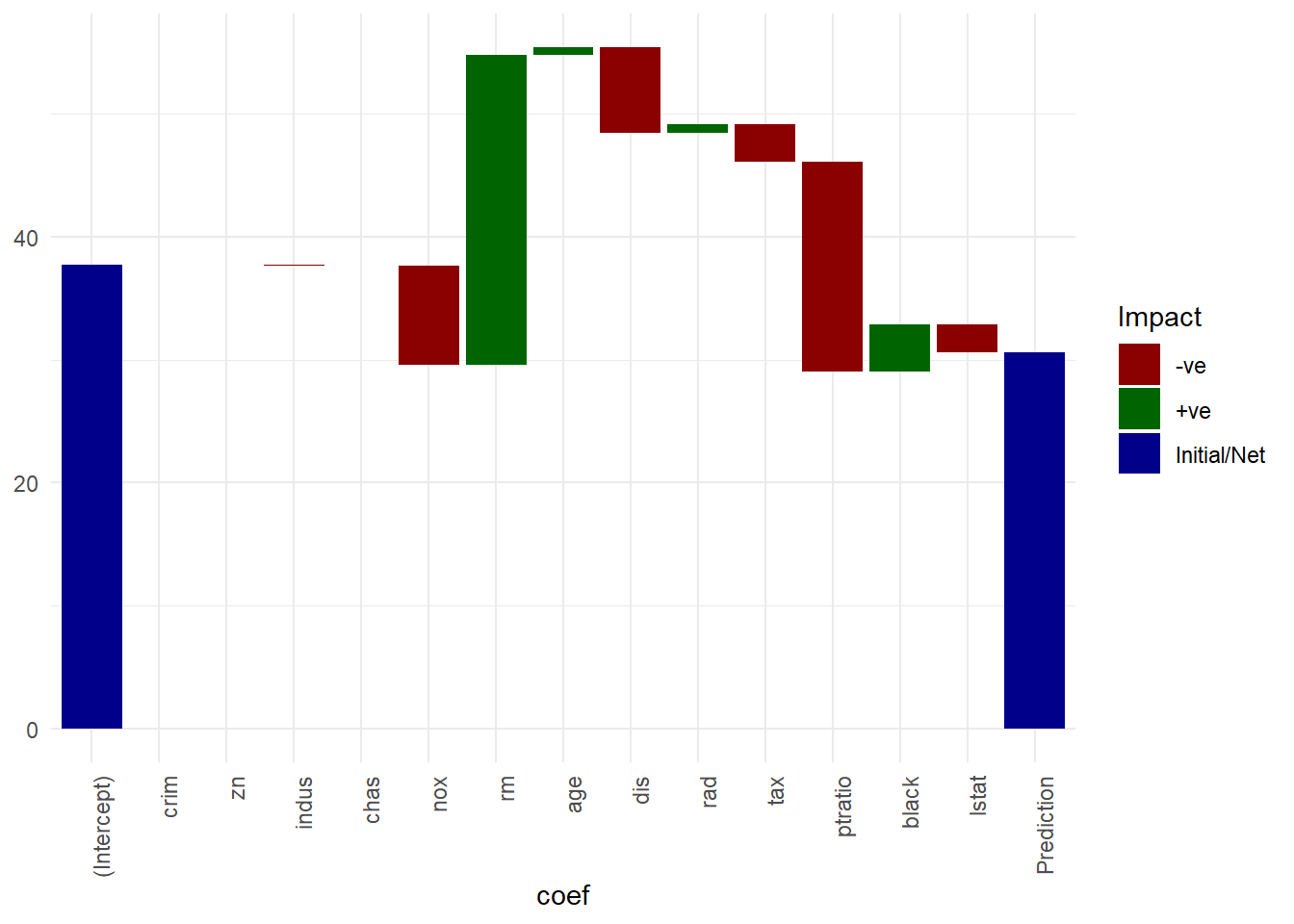
cont_prop <- featcont/pred
plot_data <- data.frame(coef = names(cont_prop),
cont_prop = cont_prop,
row.names = NULL)
plot_data <- plot_data[-nrow(plot_data),]
plot_data <- plot_data[order(plot_data$cont_prop, decreasing = FALSE),]
plot_data$coef <- factor(plot_data$coef, levels = plot_data$coef)
p<-ggplot(data=plot_data, aes(x=coef, y = cont_prop)) +
geom_bar(stat="identity", fill = "darkblue") +
coord_flip() +
theme_minimal() +
xlab("Features") +
ggtitle("Feature Contributions")
p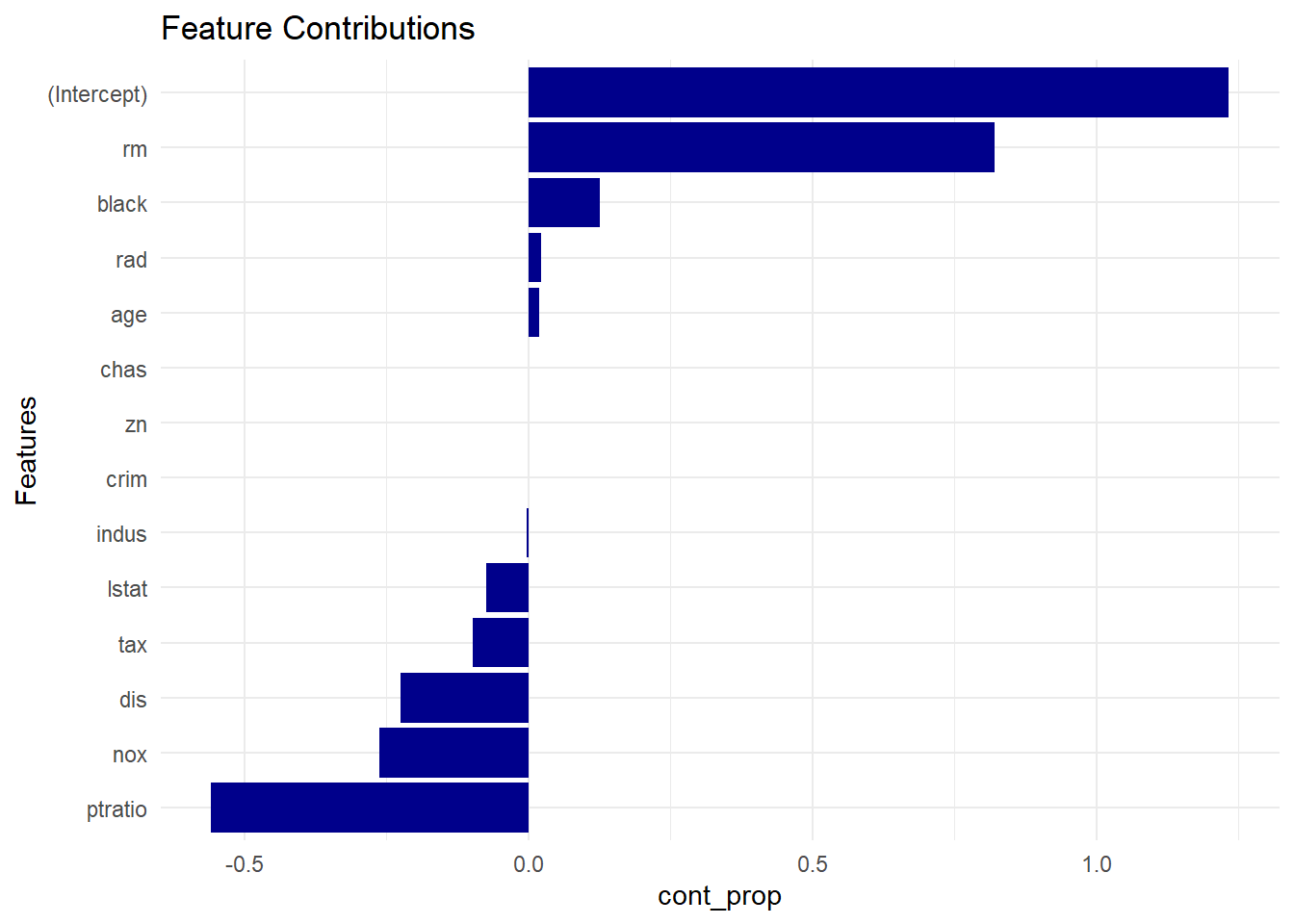
Classification
Preparation
library(kernlab)
##
## Attaching package: 'kernlab'
## The following object is masked from 'package:ggplot2':
##
## alpha
library(caret)
library(magrittr)
library(ggplot2)
data(spam)
set.seed(123)
# mean centering
s2 <- preProcess(spam, method = "center") %>% predict(., spam)
idx <- createDataPartition(s2$type, p = 0.8, list = FALSE)
train <- s2[idx,]
test <- s2[-idx,]
mod0 <- glm(data = train, type ~., family = binomial(link = logit))
## Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
sm <- summary(mod0)
betas <- sm$coefficients[,1]
testcase <- test[1,]
pred <- predict(mod0, testcase)
# dot product between feature vector and beta
featvec <- testcase[-which(testcase %>% names == "type")] %>% as.matrix
betas2 <- betas[-1]
nm <- names(betas)
#betas2 %*% t(featvec)
# feature contributions
featcont <- betas2*featvec
featcont <- c(betas[1], featcont, pred)
names(featcont) <- c(nm, "Prediction")Waterfall chart on classification feature contributions
# waterfall chart on feature contribution
plotdata <- data.frame(coef = names(featcont), featcont = featcont, row.names = NULL)
plotdata$coef <- factor(plotdata$coef, levels = plotdata$coef)
plotdata$id <- seq_along(plotdata$coef)
plotdata$Impact <- ifelse(plotdata$featcont > 0, "+ve", "-ve")
plotdata[plotdata$coef %in% c("(Intercept)", "Prediction"), "Impact"] <- "Initial/Net"
plotdata$end <- cumsum(plotdata$featcont)
plotdata$end <- c(head(plotdata$end, -1), 0)
plotdata$start <- c(0, head(plotdata$end, -1))
plotdata <- plotdata[, c(3, 1, 4, 6, 5, 2)]
gg <- ggplot(plotdata, aes(coef, fill = Impact)) +
geom_rect(aes(x = coef,
xmin = id - 0.45,
xmax = id + 0.45,
ymin = end,
ymax = start)) +
theme_minimal() +
#scale_fill_manual(values=c("#999999", "#E69F00", "#56B4E9"))
scale_fill_manual(values=c("darkred", "darkgreen", "darkblue")) +
theme(axis.text.x=element_text(angle=90, hjust=1))
## Warning: Ignoring unknown aesthetics: x
#coord_flip()
if(sign(plotdata$end[1]) != sign(plotdata$start[nrow(plotdata)]))
gg <- gg + geom_hline(yintercept = 0)
gg